COVID-19/SARS-CoV-2 Pandemic
Posted on: Monday 6 April 2020
Author: Dr Penny Ward et al.
This article has been prepared by Dr Penny Ward FFPM, with input from Professor Tim Higenbottam PFPM, Dr Flic Gabbay FFPM, Dr Bob Holland FFPM, Dr Sue Tansey FFPM and Dr Tahir Saleem MFPM.
It is provided for information and does not constitute advice or represent official FPM views or policy.
How to cite:
Ward, P et al. (2020), ‘COVID-19/SARS-CoV-2 Pandemic’, Faculty of Pharmaceutical Medicine blog, 6 April. Available at: https://www.fpm.org.uk/blog/covid-19-sars-cov-2-pandemic/ (Accessed: <date>).
This is the first part of a two-part review.
Read part two: COVID-19 Vaccine and Antiviral Drug Development
Summary
This review addresses current knowledge concerning SARS-CoV-2 and COVID-19 disease including:
- Epidemiology of the outbreak
- SARS-CoV-2 viral life cycle and clinical features of COVID-19 Disease
- Current Treatment
An enormous effort is ongoing across the life science industry in the UK and beyond to address this pandemic. It is hoped that the insights provided in this article will excite interest and support for the challenge. Future articles will address work ongoing to develop a vaccine and effective antivirals for treatment/prophylaxis.
Introduction
At the end of the Great War, a conflict which convulsed most countries in the world over four years and killed an estimated 40 million soldiers and civilians among participating nations, the world was seized by a pandemic of Influenza, which, in the space of a year, spread to every country on earth, infected almost 500 million and killed 100 million people. Three months into the 101st anniversary year of that great pandemic, we are again witnessing a rapidly spreading, highly infectious serious acute respiratory syndrome (SARS) sweep across the world. Since first recognition in Wuhan, China in December 2019 to the time of this review, Corona Virus Disease -19 (COVID-19), caused by infection with a novel coronavirus, SARS-CoV-2, has spread to virtually all countries, infected over 900,000 individuals and caused more than 40,000 deaths. It is still increasing rapidly1. This bulletin reviews the state of understanding of the virus, the pattern of disease in infected individuals and discusses advances made and work ongoing to facilitate efficient diagnosis, prevention and treatment of this significant health threat.
The Coronaviridae
Coronavirus disease was first recognised in humans in the 1930s, with the first virus (HCoV-229E) isolated in 19652. Subsequently, three further CoVs were identified in humans (HCoV-NL63, HCoVOC43 and HCoV-HKU1). These viruses are endemic in humans and, after rhinoviruses, are an important cause of the common cold, with outbreaks occurring throughout the year, but more frequently in winter and spring in temperate climates. While adults generally experience mild cold symptoms, among individuals with asthma/COPD exacerbations may occur. Infection may be more severe in infants and young children, causing tracheolaryngobronchitis (croup), bronchitis and pneumonia3. In November 2002 an outbreak of serious acute respiratory syndrome (SARS) was found to be due to a fourth, highly pathogenic, coronavirus the SARS coronavirus. The disease originated in China and subsequently spread to Vietnam, Hong Kong, Taiwan, Singapore and Canada. Between its first and last appearance, 8096 cases were reported to the World Health Organisation (WHO) with 711 deaths, giving a case fatality rate (CFR) of 9.6%. In affected countries, outbreaks were contained by vigorous identification of cases and enforced quarantining of contacts3. In Taiwan, where an outbreak followed return of an infected traveller returning from Guandong, China, 671 cases were identified and 131,132 people were quarantined4. The last case from this episode was reported in July 2003, although three small laboratory associated outbreaks have occurred since then involving 11 patients. The causative virus, the betacoronavirus, SARS CoV, has not been circulating in humans since. A fifth coronavirus was identified in 2012 the UK following hospital admission of a patient with a SARS like illness. Middle East Respiratory Syndrome (MERS) CoV has resulted in a limited number of outbreaks, mostly in Saudi Arabia and other middle eastern countries. Human to human transmission of this disease has, to date, been limited to close contacts of affected cases in households or healthcare settings. The CFR in this disease exceeds 30% 3.
Virus Structure and Replication:
Human coronavirus particles are generally spherical, 120-160nm diameter and typically decorated with large (~20 nm), club- or petal-shaped surface projections (the “peplomers” or “spikes”), which give an image resembling the solar corona on electron micrographs of infected tissues and hence to the family name (Figure 1).3
Receptor Binding
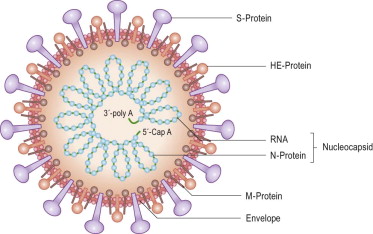
Coronaviruses have a large single positive RNA stranded genome of 28-32 kilobase size (making it the largest RNA genome of the RNA virus family) enclosed in a nucleocapsid of helical symmetry (Figure 2). These viruses infect human cells via S (spike) protein binding to receptors on host cells, followed by release of viral RNA into the cell cytoplasm. Various host receptors have been associated with the different human coronaviruses so far described: the host receptor for HCoV-229E is aminopeptidase N while HCoV-OC43 uses 2,3 or 2,6 alpha sialic acid receptors (as does influenza virus). The S protein of SARS CoV binds to angiotensin converting enzyme 2 (ACE2) and of MERS CoV to dipeptidyl peptidase 4 (DPP4)5. SARS-CoV-2 has also been demonstrated to bind to ACE2, a transmembrane receptor which is widely expressed in lung, heart, kidney and gastrointestinal tissue6.
Intracellular replication
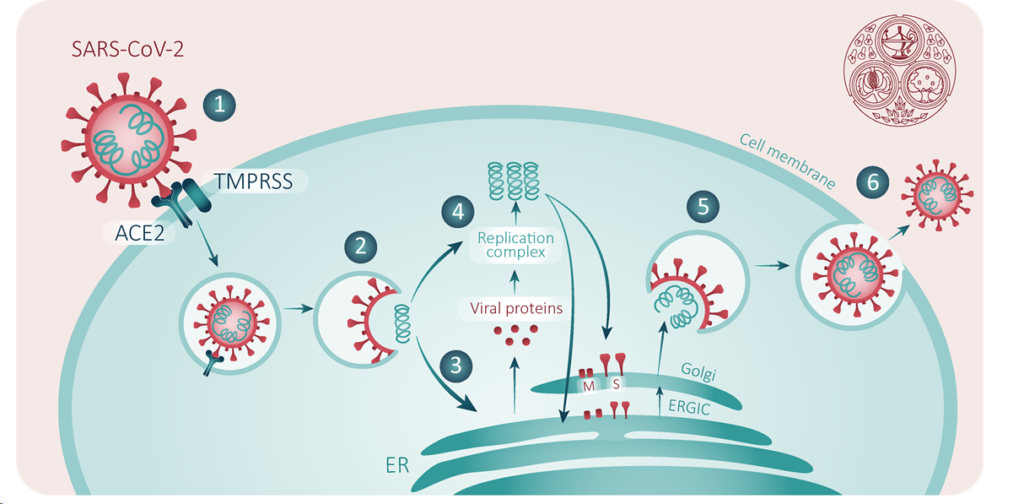
Following receptor binding, the virus must then access the cell cytoplasm. This is assisted by acid-dependent proteolytic cleavage of S protein by a cathepsin, TMPRRS2 or another protease, followed by fusion of the viral and cellular membranes. S protein cleavage occurs at two sites within the S2 portion of the protein, with the first cleavage important for separating the receptor binding domain (RBD) and fusion domains of the S protein and the second for exposing the fusion peptide (cleavage at S2′). Fusion generally occurs within acidified endosomes, but some coronaviruses can fuse at the plasma membrane. Following this, the viral replicase gene, which consists of two open reading frames (ORFs) coding for two polyproteins pp1a and pp1ab. These polyproteins contain all the non structural proteins (nsps) of the virus which are essential for intracellular replication. The polyproteins are cleaved to form individual nsps by proteases, including one or two papain-like proteases (PLpro) (depending on the coronavirus sp) and a serine type protease, the Main (M)protease. Many of these nsps then assemble into the replicase-transcriptase complex (RTC) which replicates the viral RNA. Following replication and viral RNA synthesis, the viral structural proteins, S, E, and M are translated and inserted into the endoplasmic reticulum (ER). These proteins move into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) where viral genomes encapsidated by N protein bud into membranes of the ERGIC containing viral structural proteins, forming mature virions.
Progeny virion assembly and budding
The M protein directs most protein-protein interactions required for assembly of coronaviruses but is not enough, alone, for virion formation. When M protein is expressed along with E protein, virus like particles (VLPs) are formed, suggesting these two proteins function together to produce coronavirus envelopes. N protein enhances VLP formation, suggesting that fusion of encapsidated genomes into the ERGIC enhances particle envelopment. The S protein is incorporated into virions at this step but is not required for assembly. The ability of the S protein to traffic to the ERGIC and interact with the M protein is critical for its incorporation into virions. Following assembly, progeny virions are transported to the cell surface in vesicles to be released by exocytosis (Figure 2).
Figure 2: Replication cycle of SARS-CoV-2
Route of Infection
The human coronaviruses may be spread via aerosol infection of the respiratory tract following inhalation of aerosol particles in air. Large particle sizes contaminate the upper airways, but particles of <5 microns size can move into the lower respiratory tract. Coughing, spitting, sneezing and talking generate aerosols made up of mucus droplets containing the virus. In a study investigating cough aerosols, the Edmonton group demonstrated that cough aerosols are composed of droplets ranging from 0.1 to 900 microns size, of which 97 per cent were less than 1 micron in size, and 99 per cent less than 10 microns. While larger particles will tend to settle quickly on nearby surfaces, particles less than 2.5 microns size remain airborne for a longer time. Thus, most of the infectious particles produced by coughing will remain airborne and can be inhaled into the lungs8. Coronaviruses are relatively stable on a variety of surfaces and thus can also spread via fomite transfection by hand contact with virus particles on surfaces contaminated by respiratory secretions and subsequent touching the mucous membranes of the face (eyes, mouth, nose). Human coronaviruses can be detected in faeces, and transmission via the faeco-oral route is also possible9.
Given these potential routes of infection, masking patients to provide a barrier to the coughing of aerosols into surrounding air, isolation of cases away from others, strict personal hygiene with frequent hand washing, regular cleaning of potentially contaminated surfaces and protection against aerosol inhalation by attendants by use of respiratory protection equipment are all imperative to prevent transmission.
Pattern of Illness post infection
Lauer and colleagues estimated the incubation period following initial exposure reporting a median incubation period (first exposure to onset of symptoms) of 5 days (range ~2 to ~14 days)10.
Asymptomatic Infection
The proportion of infected individuals that do not develop symptoms is unknown, as the extent of population screening has been limited. Nishiura et al investigated infection rates among a group of 565 Japanese citizens returning from China in February 2020. Of the 565 returning subjects, 14(2.4%) had a positive PCR test for SARS-CoV-2 of which 5 (41.6%: 95% confidence interval (CI): 16.7%, 66.7%) were asymptomatic at the time of testing11. Mizumoto and colleagues investigated an outbreak of SARS-CoV-2 aboard a cruise ship, the Diamond Princess. A total of 634/3063 (20.7%) subjects tested were PCR positive for SARS-CoV-2 infection. Of these 320 (50.5%) were asymptomatic at the time of testing. As testing was conducted over a 14 day period and initially focused on symptomatic cases, the authors modelled these data to reflect variability in latent period, which results in a lower estimate for the proportion of infections that remain asymptomatic – in this case 17.9% (95%CrI: 15.5–20.2%)12. Transmission of infection from asymptomatic carriers has been documented: ergo the lack of understanding of the proportion of infections which remain asymptomatic is urgently needed, both to better understand the impact of control measures as well as to correctly ascertain risk of death following infection14. The broader testing strategy pursued in Germany is thought to be one reason for the low apparent case fatality rate (~0.7%) there compared to other EU countries such as the UK (5.2%), Italy (10.6%) and Spain (7.6%), where testing is restricted to hospitalized patients.
Symptomatic Illness
Symptomatic illness is associated with fever and persistent cough with some subjects complaining of fatigue, sore throat, headache and diarrhoea. Individuals may become rapidly short of breath with increasing difficulty breathing associated with declining blood oxygenation.
Reports from China and elsewhere suggest that males may be affected more severely than females, however most patients (80%) have relatively mild, influenza-like symptoms and make a complete recovery within 10 days13.
A small proportion of predominantly older patients and those with pre-existing chronic diseases experience progressively more severe symptoms, with CT evidence of pneumonia and biochemical evidence of an exaggerated immune response potentially driven by multiple cytokines, IL-6, IL-8, IL-10, Interferon γ, and TNF-alpha which is often called a “cytokine storm”14,15. Activation of complement C3a and C5a may initiate cytokine storm and can cause thrombotic microangiopathy. It is known to be involved in many different causes of acute lung injury (ALI) and adult respiratory distress syndrome (ARDS) following viral infections16.
Children
Based on limited evidence, children are in the minority among the hospitalized cases in China, accounting for less than 1% of affected cases13,17. Zheng et al reported a small case series of 25 children affected by COVID-19 in China18. The median age of the children was 3 years, with the most severely affected subjects being mostly in this younger age group. Presenting symptoms included fever [13 (52%)], followed by dry cough [11 (44%)], diarrhea [3 (12%)], nasal congestion [2 (8%)], dyspnea [2 (8%)], abdominal pain [2 (8%)] and vomiting [2 (8%)]. Clinical diagnoses in affected cases were upper respiratory infection in 8 cases, mild pneumonia in 15 cases and critical pneumonia in 2 cases. Both critical cases had congenital heart disease and one required renal dialysis. Among the children with pneumonia, the majority of those in the younger age range had bilateral pulmonary involvement on CT. The two critical cases were thought to have evidence of secondary bacterial infections contributing to their illness. None of the patients in these case series died. Dong and colleagues recently described the epidemiology of COVID-19 in 2143 children, including both confirmed and suspected cases, in China19. Children under the age of 5 accounted for 1/3 of the cases, and disease in this age group, particularly in infants <1, was more likely to be severe and critical than in older children. The proportion of severe and critical cases was 10.6 %, 7.3%, 4.2%, 4.1% and 3.0% for the age group of <1, 1-5, 6-10, 11-15 and ≥16 years, respectively.
Pregnant Women
A small case series describing the outcome of pneumonia among 15 pregnant women in China suggested that disease was relatively mild and uncomplicated in these subjects, with normal delivery and perinatal outcomes reported20. Schwartz has summarised information from this and additional case series from China21. Maternal-foetal transmission of viral diseases is mostly (except for herpes simplex) via the hematogenous route, in which the virus circulating in the maternal blood stream enters the placenta, reaches the chorionic villous tree and foetal blood vessels, and is transmitted to the foetus. This mechanism of transmission has been shown not to occur with infection of pregnant women SARS-CoV and MERS-CoV, although the clinical infections caused by these coronaviruses has resulted in severe maternal pneumonia, maternal deaths and early pregnancy losses. In the cumulated experience from published reports of 38 pregnant women with COVID-19, of whom 37 had rt-PCR-confirmed SARS-CoV-2 infection, there were no cases of either severe pneumonia or maternal deaths. Although some subjects had additional co-morbid conditions, some of which were obstetric, these did not result in life-threatening maternal SARS-CoV 2 disease.
Transmissibility
The transmissibility of infection and illness is a critical component of epidemic projections and health service planning. Transmissibility is usually described by the calculation of the reproductive number R0, representing the average number of new infections generated by an infectious person in a totally naïve population. The complete calculation of this is hampered by our non-understanding of the portion of infections which remain asymptomatic. However, based on the incidence of transmission from identified symptomatic cases to contacts, R0 for SARS-CoV-2 ranges from 1.4 to 322. This is higher than the estimated R0 for 1918 pandemic influenza23. Thus, this epidemic, which appears to be highly transmissible, is likely to have a very significant impact on the health of populations worldwide.
Diagnosis
Nucleic Acid Amplification Tests (PCR)
One of the most impressive elements of the response of the global healthcare community to the Covid 19 outbreak has been the extremely rapid development and deployment of highly sensitive and specific tests to detect the SARS-Cov-2 virus. The first patient appears to have been hospitalised in Wuhan on 12th December and by the 2nd February a comprehensive description of the genome of the virus was published24. Reverse transcriptase polymerase chain reaction (RT PCR) reagents have been developed to detect the virus and distinguish it from other related coronaviruses and other common viral causes of cough and fever. While robust and well-established RT-PCR technology has allowed clinical laboratories with PCR machines to develop testing capability, the methodology is relatively low throughput and has been delivered through centralised services and validated molecular pathology laboratories. Samples need to be collected (throat and nasal swabs), couriered to the laboratory, tested, the results reviewed and provided to the patient/practitioner ordering the test. Although the test itself takes only a few hours, results can take several days to be delivered. The deployment of RT-PCR tests has been rather variable internationally, both in terms of the numbers of operational laboratories and in terms of the availability of testing kits. In the UK currently capacity is limited and hovering at around 5000-8000 subjects tested per day as of 010420, although attempts are being made to increase this to a rate of 25000 per day. The life sciences community is working to develop variants of the RT PCR test with higher throughput and with faster turnaround times; examples include droplet digital PCR, and of particular interest to clinicians, near patient automated PCR devices. As of 24th March more than fifty commercially available tests (nearly all PCR based) were available across the globe claiming some sort of regulatory approval (CE-mark, FDA-EAU, CLIA lab) including several near patient devices and kits.
RT-PCR tests are extremely sensitive and specific and therefore provide a “gold standard” for diagnosis of Covid 19 by the detection of SARS-Cov-2 genomic material. However, limitations of this method are that while it confirms the presence of viral RNA, it does not provide information on the continued presence of infectivity. This is an important consideration given the occasional report of a positive PCR test following previous negative assays in recovering patients. In addition, a PCR test does not detect prior infection or immunity to future infection.
Rapid Diagnostic Tests
Other approaches are now in rapid development. Alternative means to detect the virus include detection of viral proteins (immunoassays and analogous methods) and non-PCR based detection of viral genome. None of these tests are currently available clinically but they do eventually offer the possibility of rapid, close to patient testing. The UK Government has established a process for the evaluation of such tests: https://www.gov.uk/government/publications/covid-19-evaluation-of-commercially-available-diagnostic-products/guidance-for-industry-on-evaluations-of-diagnostic-products-for-coronavirus-covid-19. and is providing grants to companies developing such technologies. The development of rapid diagnostic tests will need to address the question of sensitivity – rapid bedside testing for influenza virus has been used for screening purposes for several years but the high false negative rate of these tests hampered patient care and limited public health disease control measures during the 2009-2010 pandemic, resulting in reclassification of these from Class I tests (tests with low risk to public health requiring limited monitoring) to Class II devices in the USA24. Class II devices require special controls, examples of which include performance standards, post-market surveillance, patient registries, guidelines, design controls, and other appropriate actions to mitigate risk.
Antibody Testing
The detection of an immune response to Covid 19 (serology) allows an understanding of who has been exposed (and been infected) and the degree to which individuals may have some immunity to future infections; this also facilitates better understanding of the spread of infection (as it has potential to identify asymptomatic infections). In similar viruses, the Spike (S) protein used to bind to host cells is immunogenic and the Receptor Binding Domain is the target of neutralising antibodies25. However, there are a number of important questions yet to be answered including:
- What is the cross reactivity of immunoglobulins produced after infections with other coronaviruses (specificity)?
- What proportion of infected individuals make antibodies to SARS-CoV-2?
- What magnitude of antibody response confers immunity?
- How long does this immunity last?
- Do previous Corona virus infections influence the ability to generate antibodies to SARS-CoV-2?
- Do previous Corona virus infections provide some cross protection from SARS-CoV-2?
This is a very exciting (if early) area of research and tremendous progress has been made, for example in the invention of detection methodologies for antibodies specific to SARS-CoV-226. However, it is important to remember that these tests are not suitable for acute diagnosis as it may take seven to ten days to seroconvert; equally, currently available assays may detect antibody but may not provide evidence of immunity. Therefore, the full clinical utility of such tests remains to be established by further clinical research.
Viral and Antibody Kinetics in COVID-19
Limited information from case assessments investigating viral load kinetics have suggested that viral load increases post exposure reaching a peak at or shortly following onset of symptoms27,28. More severe illness is associated with higher viral load (1 log order higher) than in patients with milder disease. Viral load generally declines over a 10-15 day period post symptom onset, but may be sustained, and at higher level, in subjects developing severe pneumonia/ARDS29. The pattern of viral load dynamics is similar to influenza infection, implying that antiviral therapy given earlier in the disease course may prevent later complicated disease. This has implications for the design of clinical trials with antiviral therapies.
A recent series investigating antibody to SARS-CoV-2 viral proteins demonstrated that rising antibody levels can be detected from approx. day 7 of illness with most patients seroconverting within 14-21 days of onset. IgG and IgM antibody levels against the SARS-CoV-2 internal nucleoprotein and the surface spike receptor binding domain correlated with neutralising activity30.
Current Treatment
There are, currently, no specific antiviral therapies available for the treatment of COVID-19. Treatment of disease is currently supportive management based on first principles of respiratory support and management of complications.
Supportive Treatment
In the absence of specific interventions or availability of convalescent serum, management of COVID-19 is limited to the alleviation of symptoms, oxygen, continuous positive airways pressure and in more severe cases with ARDS and/or impending respiratory failure ventilatory support or exogenous corporeal membrane oxygen therapy. Approx 15-33% of COVID-19 affected hospitalised cases develop acute respiratory distress syndrome (ARDS). Factors that increase the risk of developing ARDS and death (which may occur in 40-50% of affected cases) include older age, neutrophilia, elevated lactate dehydrogenase level, CRP and D-dimer level and reduced platelet count.
Management of ARDS
Low tidal volume, plateau-pressure-limited mechanical ventilation is the primary treatment that has been shown to reduce mortality from ARDS. Tracheal intubation before the start of ventilatory support is a high risk, critical procedure in patients with COVID-19. Healthcare staff must apply the SAS principles: Safe – for staff and patient; Accurate – avoiding unreliable, unfamiliar or repeated techniques; Swift – timely, without rush or delay. Low tidal volume, plateau-pressure-limited mechanical ventilation is the primary treatment that has been shown to reduce mortality. Tidal volumes should be targeted to a lung-protective range (4-6 ml/kg ideal body weight). Informal reports from Italy and Singapore suggest that the driving pressures required are not very high, that patients require positive end pressures (PEEP) and respond well to prone ventilation. This suggests that the primary pulmonary pathology is likely to be small airway closure and atelectasis, rather than reduced lung compliance. Complications of ventilation include pneumothorax, ventilator-associated pneumonia, multiple organ failure, and, in the longer term, pulmonary fibrosis with prolonged respiratory failure.
Extrapulmonary Manifestations
Hepatic dysfunction (elevated transaminases) has been reported in 14% to 53% of patients in case series, more commonly in patients with severe disease. Evidence suggests that clinically significant liver injury is uncommon.
Cardiac injury is reported in 7% to 13% of patients in case series, with higher prevalence in patients who are severely or critically ill. Myocardial injury has been reported in > 25% of patients who are critically ill, and presents in two ways: acute myocardial injury and dysfunction on presentation or myocardial injury that develops as the severity of illness worsens. Arrhythmias have been reported in 16% of patients in case series. Fulminant myocarditis has been reported while cardiomyopathy has been reported in 33% of critically ill patients. It is unknown whether it is a direct cardiac complication of COVID-19 or due to overwhelming clinical illness. The long term consequence of infection on cardiovascular health is unknown.
Acute kidney injury (AKI) has been reported in 3-8% of cases, mostly those with other complications requiring ICU support: early recognition is important as AKI is associated with inpatient mortality >20%.
Disseminated intravascular coagulation is a feature in 71% of patient dying from COVID-19. It is frequently associated with other complicated illness in ICU.
As with any viral infection affecting the lungs, secondary bacterial infection and sepsis may further complicate the course of illness31. Elevated procalcitonin may enable earlier recognition of this potential and appropriate, early, use of antibiotics32.
Convalescent Serum
Convalescent serum is a strategy which has been exploited previously in novel infectious disease and is being explored in clinical trials. This approach has been reported to be effective in patients with SARS and MERS33,34. In a retrospective analysis use of convalescent plasma in SARS patients in Hong Kong, treatment was associated with a significantly reduced mortality and higher clinical success rate (defined as the proportion leaving hospital by day 22) compared to those receiving only corticosteroid therapy35. Patients receiving convalescent plasma after day 16 fared no better than those receiving steroid alone35. With rising numbers of COVID-19 patients, consideration should be given to preparation of convalescent plasma/serum from affected subjects for use in patients requiring hospitalization for more severe disease.
Conclusions
The emergence and rapid spread of SARS-CoV-2 represents a major emerging health threat to countries and populations around the world. Currently countries are enforcing public health measures designed to reduce spread of the disease within the population. Evidence from countries affected early in the outbreak suggests that these will successfully control spread, but comes with significant population disruption and potential for adverse effects on the world economy. An enormous effort is ongoing to understand the disease pathogenesis and to convert that understanding to the rapid scale up and production of therapeutic and prophylactic approaches to protect exposed populations. A range of promising avenues has been identified and will be discussed in the next chapters. In the interim, convalescent plasma/serum may offer an early approach to the specific management of disease in patients requiring hospitalization.
References
- WHO Situation Report 1 April 2020; https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200329-sitrep-69-covid-19.pdf?sfvrsn=8d6620fa_4 accessed 2 April 2020.
- Kahn, J, McIntosh, K. “History and recent advances in coronavirus discovery”, Pediatric Infectious Disease Journal 2005; 24: s223–s227
- Korsman S et al. Human Coronaviruses. Virology 2012. Pub. Churchill Livingstone
- Lee ML et al. Use of quarantine to prevent transmission of Severe Acute Respiratory Syndrome – Taiwan, 2003. MMWR 2003. 52: 680-83
- Widagdo W et al. Host Determinants of MERS-CoV Transmission and Pathogenesis. Viruses. 2019; 11: 280
- Letko, M. et al. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5, 562–569
- Fehr AR and Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol Biol. 2015; 1282: 1–23.
- Zayas G et al. Cough aerosol in healthy participants: fundamental knowledge to optimize droplet-spread infectious respiratory disease management. BMC Pulmonary Medicine 2012, 12:11
- Zhou J et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017; 3: eaao4966
- Lauer S et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020. DOI: 10.7326/M20-0504
- Nishiura H et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int J Infect Dis doi: 10.1016/j.ijid.2020.03.020
- Mizumoto K and Chowell G. Transmission potential of the novel coronavirus (COVID-19) onboard the diamond Princess Cruises Ship 2020. Infectious Disease Modelling 2020. 5: 264-270
- The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. Vital Surveillances: The Epidemiological Characteristics of an Outbreak of 2019 Novel Coronavirus Diseases (COVID-19) — China, 2020. China CDC Weekly 2020; 2: 113-122
- Spadaro S et al. Biomarkers for Acute Respiratory Distress syndrome and prospects for personalised medicine. Journal of Inflammation 2019; 16:1
- Gao Y et al. Diagnostic Utility of Clinical Laboratory Data Determinations for Patients with the Severe COVID-19. J Med Virol 2020. https://doi.org/10.1002/jmv.25770
- Gralinski LE et al. Complement Activation Contributes to Severe Acute Respiratory Syndrome Coronavirus Pathogenesis. mBio 2018; 9: 1753-18. https://doi org/10.1128/mBio.01753-18.
- Liu H et al. Clinical and CT imaging features of the COVID-19 pneumonia: Focus on pregnant women and children. J Infect 2020. https://doi.org/10.1016/j.jinf.2020.03.007
- Zheng F et al. Clinical Characteristics of Children with Coronavirus Disease 2019 in
Hubei, China. Current Medical Science 2020; 40: 1-6 - Dong Y et al. Epidemiological Characteristics of 2143 Pediatric Patients With 2019 Coronavirus Disease in China. Pediatrics. 2020; doi: 10.1542/peds.2020-0702
- LIU D et al. Pregnancy and Perinatal Outcomes of Women With Coronavirus Disease (COVID-19) Pneumonia: A Preliminary Analysis. Am J Roentgenol. 2020:1-6. doi: 10.2214/AJR.20.23072.
- Schwartz DA. An Analysis of 38 Pregnant Women with COVID-19, Their Newborn Infants, and Maternal-Fetal Transmission of SARS-CoV-2: Maternal Coronavirus Infections and Pregnancy Outcomes. Archives of Pathology & Laboratory Medicine 2020. In-Press.
- Liu Y et al. The reproductive number of COVID-19 is higher compared to SARS coronavirus. Journal of Travel Medicine 2020. 27; https://doi.org/10.1093/jtm/taaa021
- Biggerstaff M et al. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis. 2014; 14: 480. doi: 10.1186/1471-2334-14-480.
- Green DA, St. George K. Rapid antigen tests for influenza: rationale and significance of the FDA reclassification. J Clin Microbiol 2018; 56:e00711-18. https://doi.org/10.1128/JCM. 00711-18.
- Wu, F et al. A new coronavirus associated with human respiratory disease in China. Nature 2020; 579: 265–269
- Amanat, F., Nguyen, T., Chromikova, V., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv preprint doi: https://doi.org/10.1101/2020.03.17.20037713.
- Pan Y et al. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 2020 https://doi.org/10.1016/S1473-3099(20)30113-4
- Zou L et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020; (published online Jan 30.) DOI:10.1056/NEJMc2001737
- Liu Y et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis 2020. DOI:https://doi.org/10.1016/S1473-3099(20)30232-2
- To K K-W et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020. https://doi.org/10.1016/S1473-3099(20)30196-1.
- https://bestpractice.bmj.com/topics/en-gb/3000168
- Wirz Y et al. Effect of procalcitonin-guided antibiotic treatment on clinical outcomes in intensive care unit patients with infection and sepsis patients: a patient-level meta-analysis of randomized trials. Critical Care 2018; 22:191
- Arabi Y, Balkhy H, Hajeer AH, et al. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. Springerplus. 2015;4:709. https://doi.org/10.1186/s40064‐015‐1490‐9
- Cheng Y, Wong R, Soo YOY, et al. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis. 2005;24: 44‐46. https://doi.org/10.1007/s10096‐004‐1271‐9
- Soo YOY et al. Retrospective comparison of convalescent plasma with continuing high‐dose methylprednisolone treatment in SARS patients. Clin Microbiol Infect. 2004; 10:676‐678.https://doi.org/10.1111/j.1469‐0691.2004.00956.
Read part two of this article
- COVID-19 Vaccine and Antiviral Drug Development
